Stitching overlapping volumes#
In the case where the same object is described two separate 3D volumes with some overlap it can be useful to merge the two volumes into one.
Obviously DVC has a role to play in this – since it can help to align one of the two images onto the other in order to have a good match. In this note we’ll see how to do this on a dataset of a sandstone acquried at the Diamond Light Source by Alexis Cartwright-Taylor and the Edinburgh Geoscience Microtomography team [*] in 2019.
There are two volumes and the overlap in mm given by the scanner is about 520 pixels. Here we will deform the “top” image to make it match the bottom image.
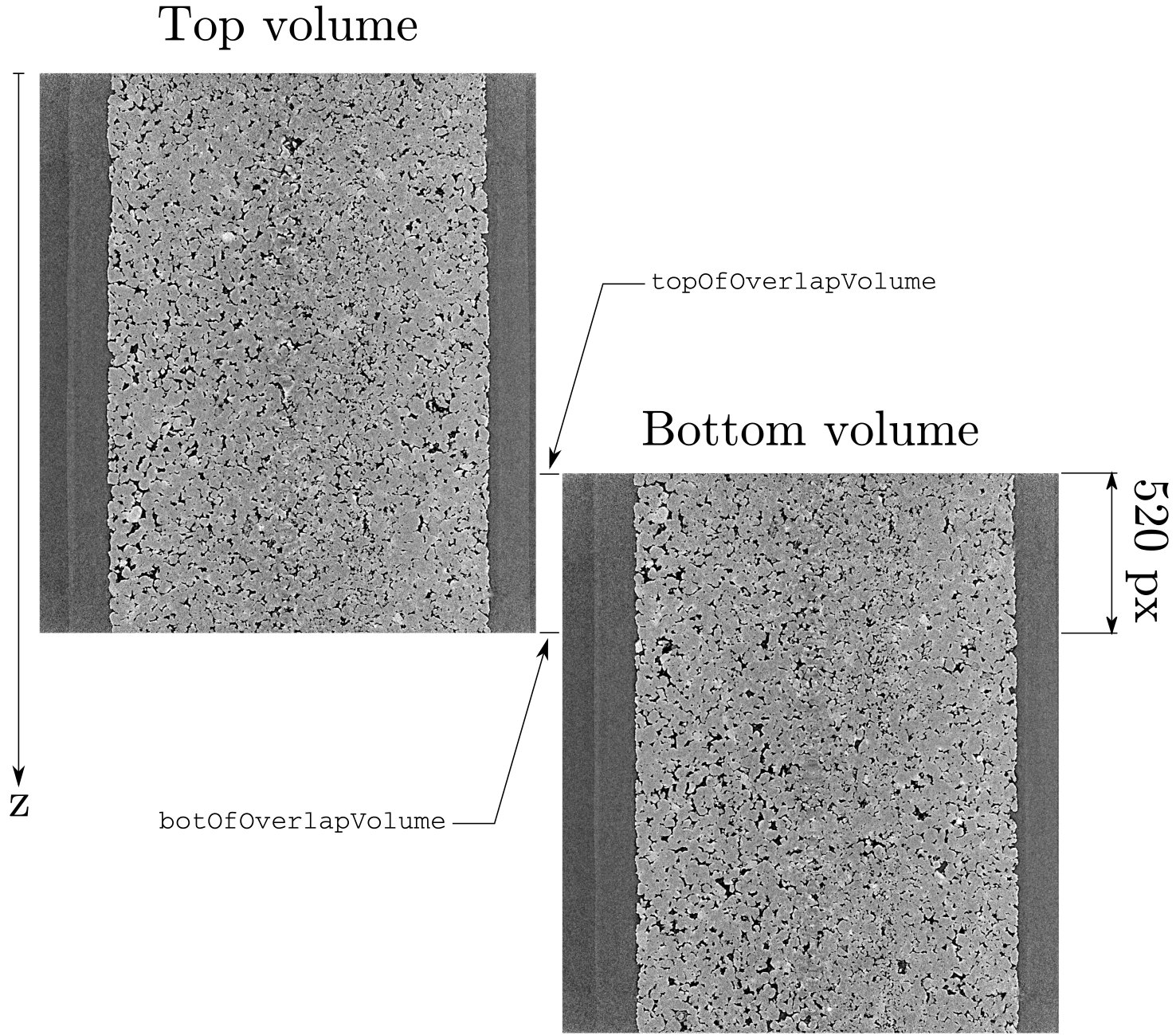
Vertical slices through top and bottom volumes#
If we try a naive approach by just assuming there is no shift between the images, we get a very poor overlap:
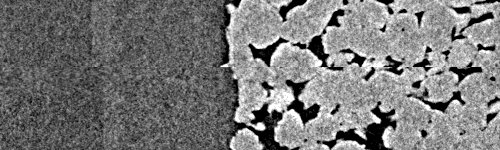
Zoom into overlap zone with naive stitching#
We will start by doing a registration between the bottom 520 pixels of the top scan and the top 520 images of the bottom scan, the measured deformation will then be applied to the top scan, using the middle of the overlap zone as the centre of application:
import tifffile
import numpy
import tifffile
import spam.DIC
import spam.deformation
# scan number
n = 0
# "Height" of overlap zone in pixels
overlap = 520
top = tifffile.imread("top/{:02d}*.tif".format(n))
bot = tifffile.imread("bot/{:02d}*.tif".format(n))
# Define the output volume -- the sum of the two heights minus overlap
merged = numpy.zeros((top.shape[0]+bot.shape[0]-overlap, bot.shape[1], bot.shape[2]), dtype='<u2')
### Perform registration, here it's pertinent to use the multiscale registration
# It might be necessary to apply an initial guess here with PhiInit
reg = spam.DIC.registerMultiscale(top[-overlap:], bot[0:overlap],
4, margin=32, verbose=True,
imShowProgress='X')
# Compute the rigid part of the measured Phi
PhiRigid = spam.deformation.computeRigidPhi(reg['Phi'])
### Apply the rigid Phi to the middle of the "top" overlap volume
topDef = spam.DIC.applyPhi(top,
Phi=PhiRigid,
# _z-centre of overlap zone_ middle-y middle-x
PhiCentre=[int(top.shape[0]-overlap/2), (top.shape[1]-1)//2, (top.shape[2]-1)//2 ])
# The z-position of the top and bottom of the overlap zone, in the coordinate system of "top"
topOfOverlap = top.shape[0]-overlap
botOfOverlap = top.shape[0]
# Let's fill in the non-overlapping top and bottom from topDef and bot:
# top
merged[0:topOfOverlap] = topDef[0:topOfOverlap]
# bottom
merged[botOfOverlap:] = bot[overlap:]
# Now on the overlap zone, take smooth average of both images:
# 1st overlap image will be 519/520 top and 1/520 bottom
# middle will be 50-50
for i in range(overlap):
merged[i+topOfOverlap] = (overlap-1-i)/(overlap-1)*topDef[i+topOfOverlap] + (i)/(overlap-1)*bot[i]
# If wanted, save result in merged folder
tifffile.imsave("merged/{:02d}-merged.tif".format(n), merged.astype('<u2'))
The result is much better:
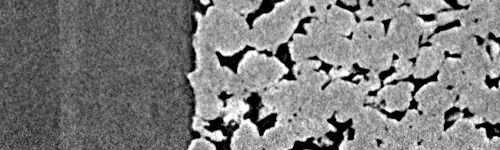
Zoom into overlap zone with DVC-deformed top and smooth merge#
